Volume 284, Number 5411 Issue of 2 Apr 1999,
pp. 80 - 81
©1999 by The American Association for the Advancement of Science.
COMPLEX SYSTEMS:
Building Working Cells 'in Silico'
Dennis Normile*Cells provide living proof of that old saw about the whole being greater than the sum of its parts. "Even if you construct a complete list of all the processes known to occur within a cell, that won't tell you how it works," says Masaru Tomita, a professor of bioinformatics at Keio University in Fujisawa, near Tokyo. But Tomita, who is a computer scientist as well as a biologist, has a scheme for exploring the effects that only emerge when those many processes interact: a simulation program that can reproduce, in simplified form, a cell's biochemical symphony.
His group's E-CELL simulation software will go on the Web for public "beta" testing this June (www.e-cell.org). Other computer models of the cell are being developed, but they often try to reproduce individual cellular processes in detail. E-CELL, in contrast, is designed to paint a broad-brush picture of the cell as a whole. Such efforts "are a next logical step" now that genome sequencing is giving biologists the complete parts lists for living things, says Peter D. Karp, a bioinformaticist at Pangea Systems, a bioinformatics software company in Menlo Park, California.
E-CELL is actually a model-building kit: a set of software tools that allows a user to specify a cell's genes, proteins, and other molecules, describe their individual interactions, and then compute how they work together as a system. It should ultimately allow investigators to conduct experiments "in silico," offering a cheap, fast way to screen drug candidates, study the effects of mutations or toxins, or simply probe the networks that govern cell behavior.
Stripped-down cell. Biochemistry simulated by E-CELL Software.
Written to run under the UNIX or Linux operating systems, the software relies on the user to input a cell's molecules, their locations and estimated concentrations within the cell, and the reaction rules that govern them. E-CELL then computes how the abundance of each substance at a particular location changes at each time increment. With a single mouse click, the user can knock out particular genes or groups of related genes, expose the cell to a foreign substance or deprive it of a nutrient, and then run the simulation again. Graphical interfaces allow the user to monitor the cell's changing chemistry.
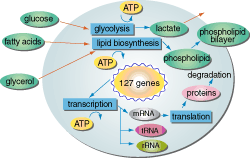
Tomita's group has used early versions of E-CELL to construct a hypothetical cell with 127 genes, which they figured was a minimal set for a self- sustaining cell in their system. Most of the genes were based on those of Mycoplasma genitalium, a microbe that has the smallest known gene set of any self-replicating organism. But the genes for some vital cellular processes still have not been identified in the mycoplasma, so the group added genes from other organisms. The virtual cell "lives," maintaining a simple, stable metabolism: It takes up glucose from the virtual culture medium, generates the enzymes and proteins to sustain internal cell processes, and exports the waste product lactate.
This bare-bones cell has already delivered one surprise. As expected, starving it of glucose causes a drop in levels of adenosine triphosphate (ATP), a key compound that provides the energy for many intracellular processes. But unexpectedly, before ATP levels drop they briefly rise. The reason, Tomita suspects, is that the early part of the ATP-producing pathway itself consumes ATP. Cutting the supply of glucose shuts down the early stages of the pathway, stopping ATP consumption there even while ATP continues to be produced from intermediary metabolites further down the pathway. Tomita thinks the effect may eventually be confirmed in living cells.
More surprises could be forthcoming when E-CELL is eventually put to work simulating whole cells of real organisms. Tomita admits that because building model cells with E-CELL depends on understanding the functions of large numbers of genes, the software is not likely to prove really useful for molecular biologists for some time. But he and his colleagues designed the program so that it should easily scale up to simulating the thousands of genes in a real cell. "Tomita and his group have done a fantastic job of engineering a 'graphical cockpit' for initializing and monitoring a whole-cell simulation," says Karp.
For greater realism on a smaller scale, users can turn to a different model-building kit: the Virtual Cell developed by physiologist Leslie Loew and computer scientist James Schaff of the University of Connecticut Health Center in Farmington. Rather than downloading software to run on their own computer, Virtual Cell users will simply run their simulation on Loew's host computer via the Internet. More important, rather than simulating an entire cell at once, as a biochemical system, Virtual Cell will eventually enable cell biologists to study how a cell's shape, volume, and other physical features affect individual biochemical processes.
Loew's team builds its Virtual Cell models using precise measurements of how molecules diffuse and react within living cells, which they make by labeling key molecules and observing them with a video microscope. The result is a computerized cell with physical properties resembling those of real cells--a framework in which users can unleash specific biochemical reactions. For example, a researcher can add a certain amount of calcium--a key intracellular messenger--and then sit back and let the Virtual Cell solve equations describing reaction and diffusion rates for each of the molecular participants affected by calcium. Then the program generates a movie of the process. "The simulations are comfortable for the biologists to use because they are based on real image data," Loew explains.
In the case of calcium, the simulation not only looked much like the calcium waves measured in actual cells--indicating that the simulation was realistic--but it also predicted the dynamics of an intermediary molecule called IP3, which cannot be monitored inside the cell itself. (Demonstrations of Virtual Cell can be accessed at www.nrcam.uchc.edu)
"These two approaches can complement each other very well," Tomita says. And both are attracting growing interest from other biologists. Tomita says that when he first starting describing his plans for E-CELL, "I was dismissed as a naïve computer scientist." Now he gets e-mail requests for information on his simulation software nearly every day. Loew, too, has found that "interest has begun to mushroom." He adds, "[Cell biologists] are getting to the point that they are realizing that without computers we are never going to be able to organize all this information."
With reporting by Elizabeth Pennisi.